Network visualization - Part 3
In the previous two posts, we discussed about IGRAPH object and how to manipulate, measure and cluster it. In this final post of network analysis series, I will focus on the network work visualization.
Network visualization are supported by two aspects — the aesthetics of network elements (aka, vertices and edges) and layout of network. There are multiple packages available for these aspects. I will focus on the basic igraph plot which is base R plot and the application of ggraph which use similar syntax comparable to ggplot2.
Aesthetics of network elements
The aesthetics of both vertices and edges can be manipulated at color, transparency. Specially for vertices, we can also manipulate its shape, size and fill. For edges, we can manipulate its width/thickness, linetype, arrow and so on. Here, use simple example “actors” to show you how to present aesthetics using igraph default plot and ggraph
Code
actors <- data.frame(
name=c("Alice", "Bob", "Cecil", "David","Esmeralda"),
age=c(48,33,45,34,21),
gender=c("F","M","F","M","F"))
relations <- data.frame(
from=c("Bob", "Cecil", "Cecil", "David","David", "Esmeralda"),
to=c("Alice", "Bob", "Alice", "Alice", "Bob", "Alice"),
same.dept=c(FALSE,FALSE,TRUE,FALSE,FALSE,TRUE),
friendship=c(4,5,5,2,1,1),
advice=c(4,5,5,4,2,3)
)
g <- graph_from_data_frame(relations, directed=TRUE, vertices=actors)Vertex aesthetics
Specify aesthetics in vertex attribute
Code
# make female and male color different
v = as_data_frame(g, what="vertice") %>% as_tibble %>%
mutate(color=case_when(gender=="F" ~ "red", gender=="M" ~ "blue"))
g = g %>% set_vertex_attr("color", value=v$color)
plot(g)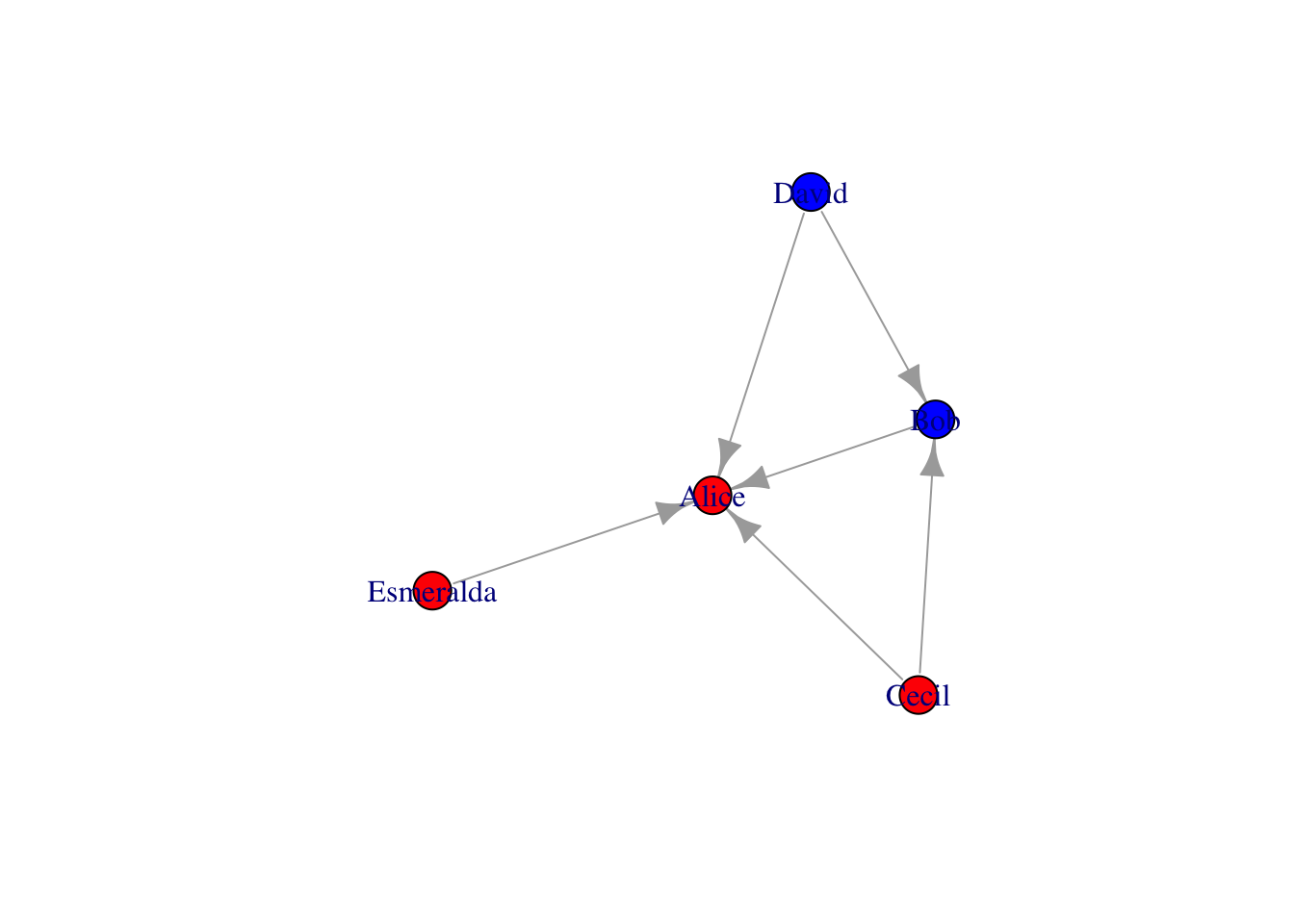
Code
# make age as size
v = v %>%
mutate(size=case_when(age < 30 ~ 10, age < 40 & age >30 ~ 20, age > 40 ~ 30))
g = g %>% set_vertex_attr("size", value=v$size)
plot(g)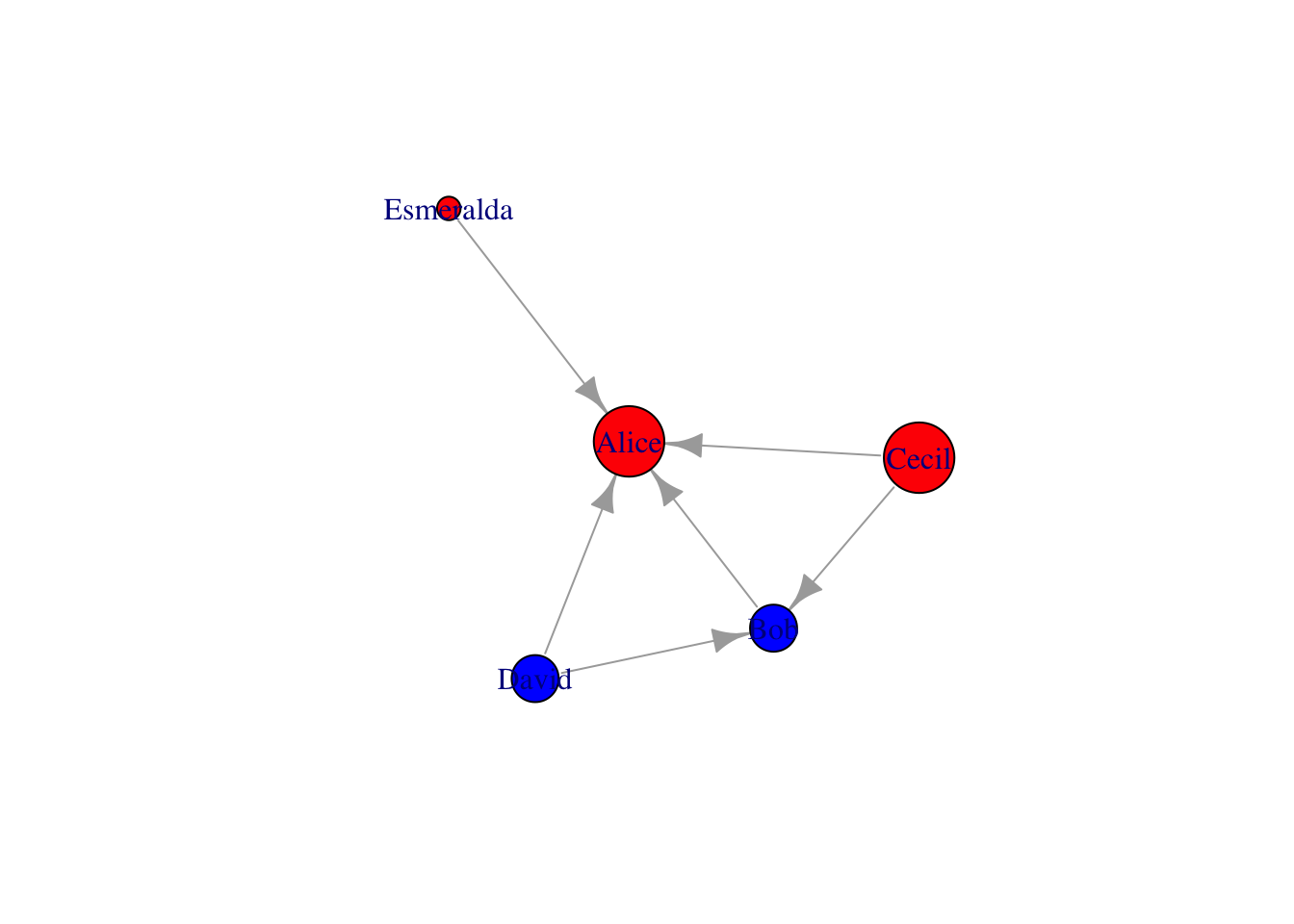
The methods mentioned above can also be done by specify in plot(). One quick example below show the shape aesthetics. Check igraph valid shape names by names(igraph:::.igraph.shapes)
Code
# make gender as shape
v = v %>%
mutate(shape=case_when(gender=="F" ~ "circle", gender=="M" ~ "rectangle"))
plot(g, vertex.shape=v$shape)
legend('topleft',legend=unique(v$gender),pch=c(21, 22),pt.bg=c("red","blue"))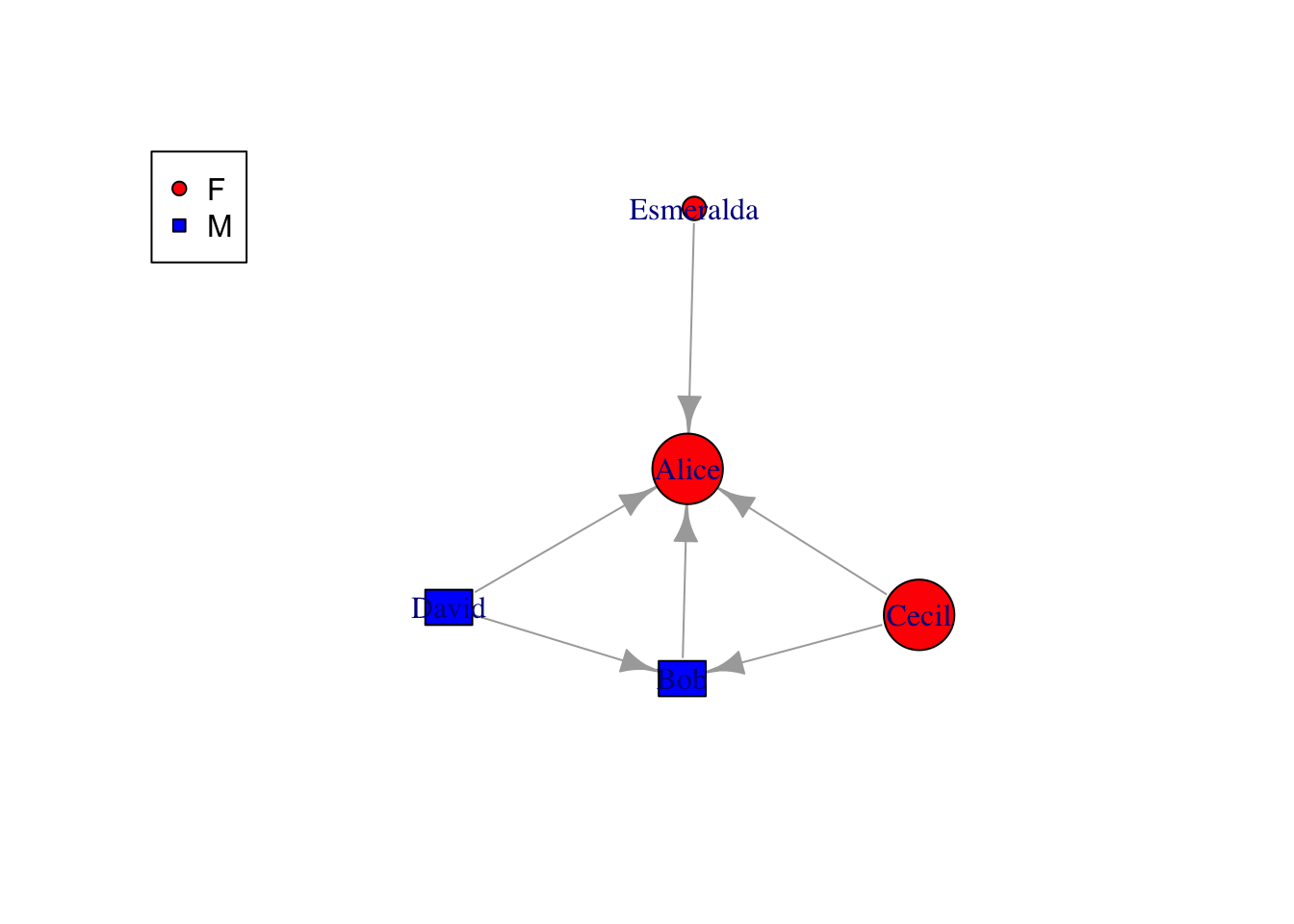
Be aware that the aesthetics specified by attributes can be overwritten by specifying in plot(). In addition, those aesthetics can also be used to apply to all vertices like plot(g, vertex.shape="rectangle"). The attributes to be manipulated in igraph (using base R) are limited. To find all the plotting attributes, try ?plot.igraph or go to https://igraph.org/r/doc/plot.common.html
We can also draw attention to certain nodes by mark.groups in plot
Code
# mark dept
g = g %>% set_vertex_attr("dept",value=c("sale","IT","sale","IT","sale")) %>%
set_edge_attr("same.dept",value=c(F,F,T,F,T,T))
v = as_data_frame(g, "vertices")
plot(g,
mark.groups=list(
unlist(v %>% filter(dept=="sale") %>% select(name)),
unlist(v %>% filter(dept=="IT") %>% select(name))
),
mark.col=c("#C5E5E7","#ECD89A"), mark.border=NA)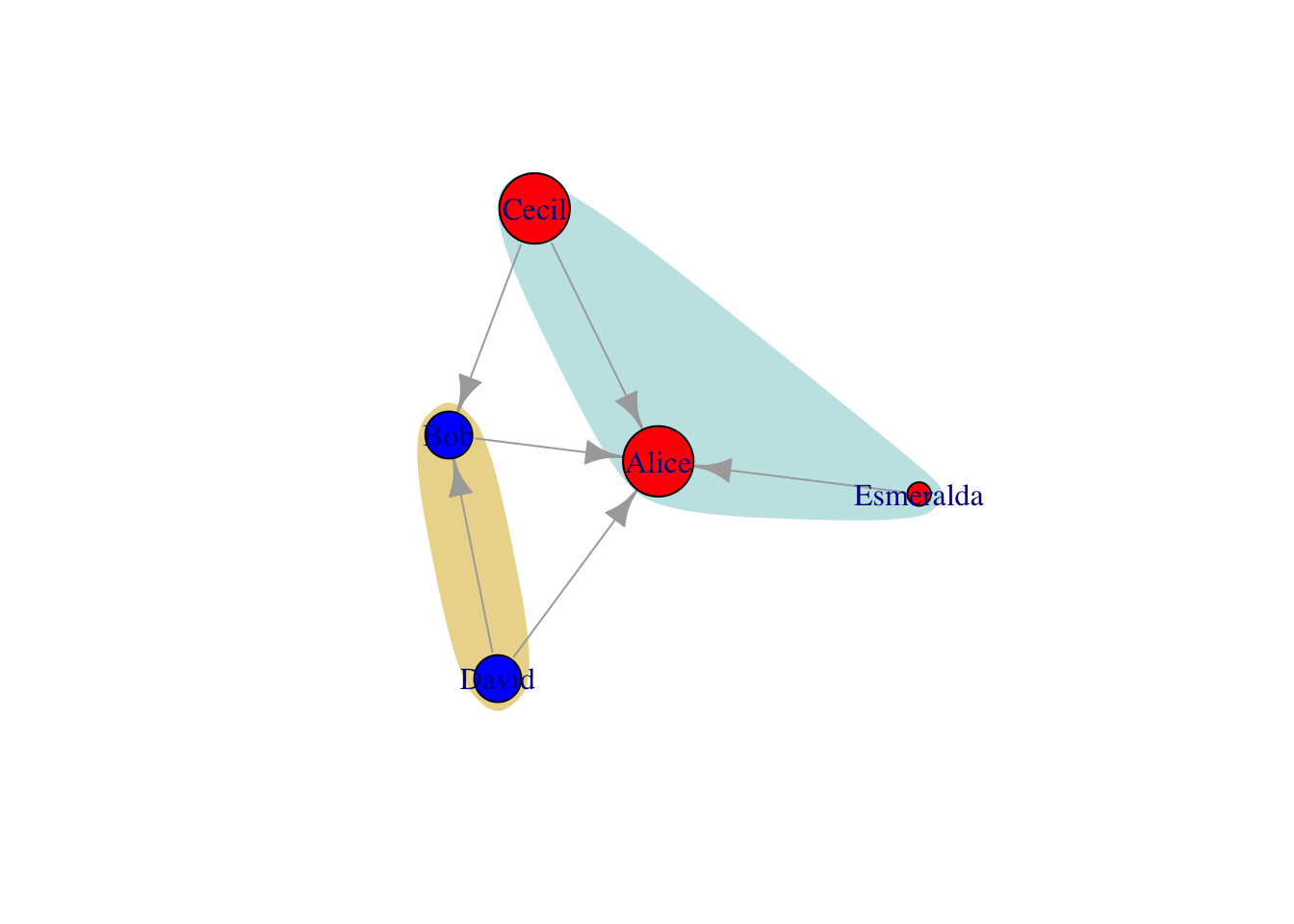
ggraph is a ggplot version of graph plotting. Using graph object as input, it can convert vertice attributes to plot attribute automatically or manually.
Code
v = v %>%
mutate(age_range=case_when(age < 30 ~ 20, age < 40 & age >30 ~ 30, age > 40 ~ 40))
g = g %>% set_vertex_attr("age_range", value=v$age_range)
ggraph(g, layout = "kk") +
geom_node_point(aes(size=age_range, color=gender), alpha=0.5) +
geom_node_text(aes(label=name)) +
geom_edge_link() +
scale_size_continuous(breaks=c(20,30,40), range = c(2, 6)) +
theme_void() 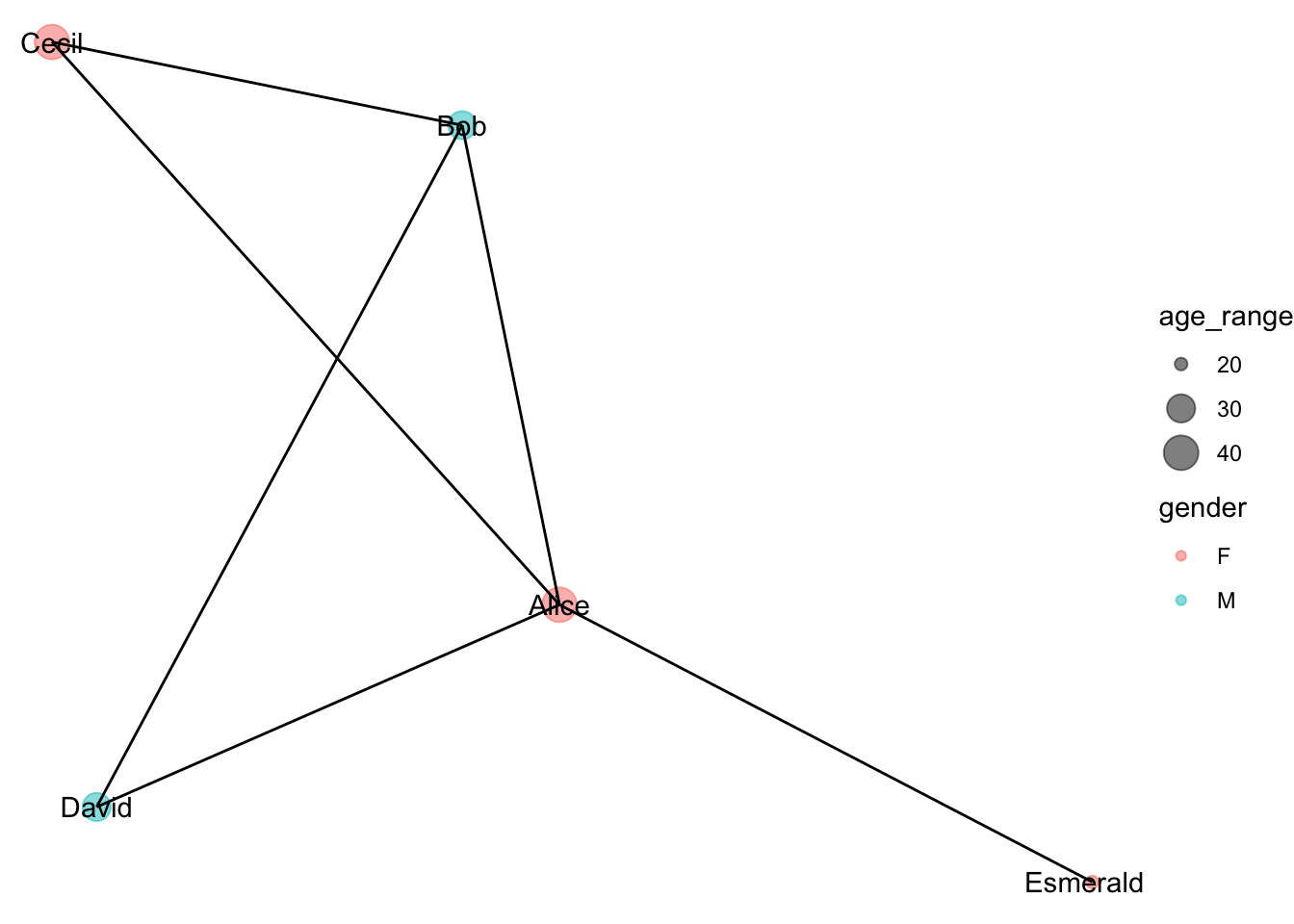
Almost all the {ggplots} theme, scale functions are available for {ggraph}. Refer to rdocumentation for more details.
Edge aesthetics
Similar to vertex aesthetics, edge plotting aesthetics can be manipulated both {igraph} default plotting and {ggraph} plotting
Code
# use linetype present whether come from same department, and line width presents friendship
e = as_data_frame(g, what="edges") %>% as_tibble %>%
mutate(width=friendship) %>%
mutate(lty=ifelse(same.dept,1,2))
plot(
g %>% set_edge_attr("width",value=e$width) %>% set_edge_attr("lty",value=e$lty),
edge.arrow.size=0.8,
edge.curved=T
)
legend("topleft", legend=unique(v$gender),pch=21,pt.bg=c("red","blue"), title="gender", box.lty=0)
legend("left",legend=unique(e$same.dept),lty=c(1,2), title = "same.dept",box.lty=0)
legend("topright", legend=sort(unique(e$friendship)), lwd=sort(unique(e$friendship)), title="friendship", box.lty=0)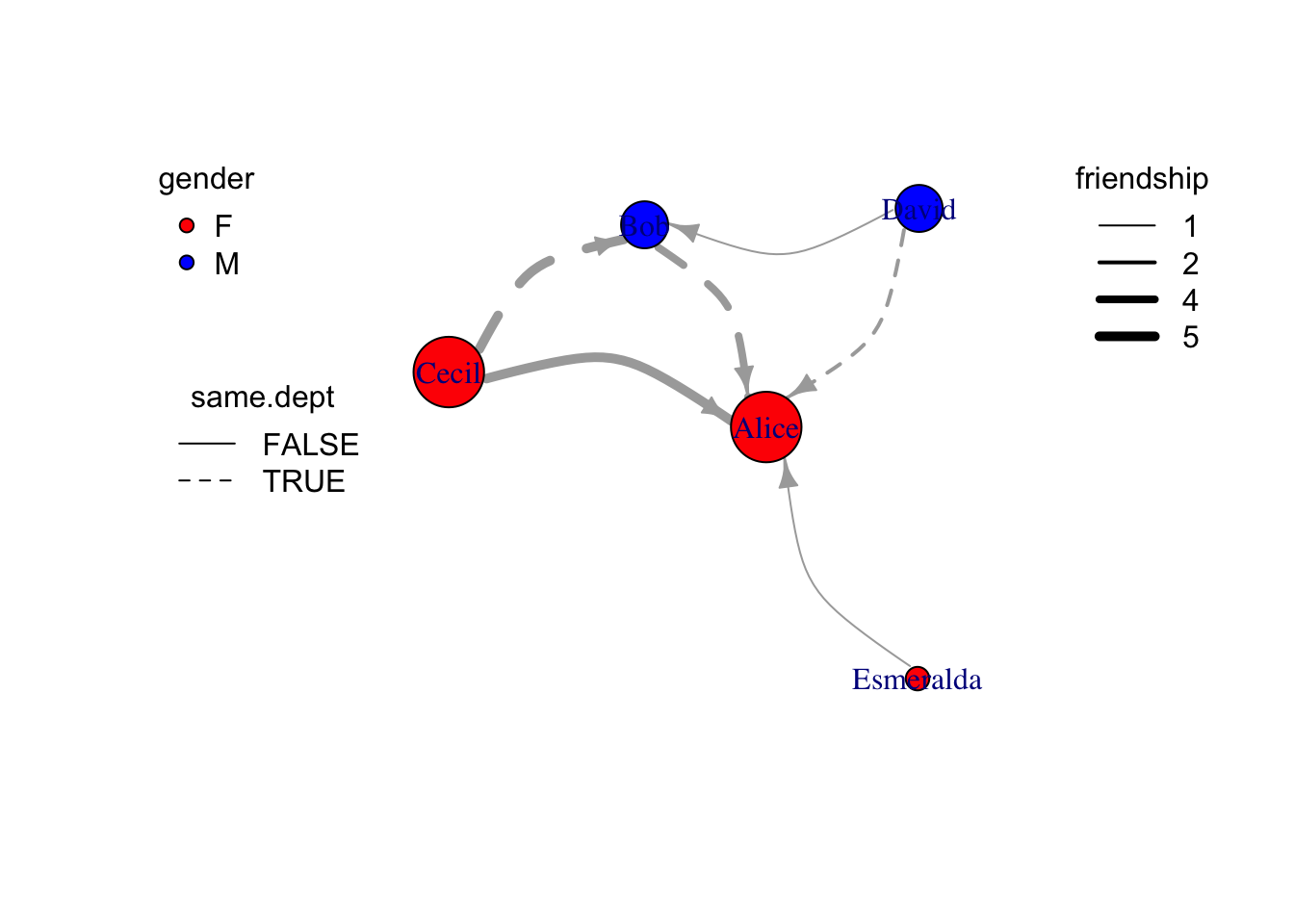
Using {ggraph} to show edges attribute is much easier.
Code
ggraph(g, layout="kk") +
geom_edge_link(aes(edge_width=friendship, edge_linetype=same.dept), arrow = arrow(angle=5, length = unit(0.3, "inches"))) +
geom_node_point(aes(color=gender), size=6) +
geom_node_text(aes(label=name), nudge_y = -0.1, nudge_x = -0.1) +
scale_edge_width(range = c(1, 2)) +
theme_void() 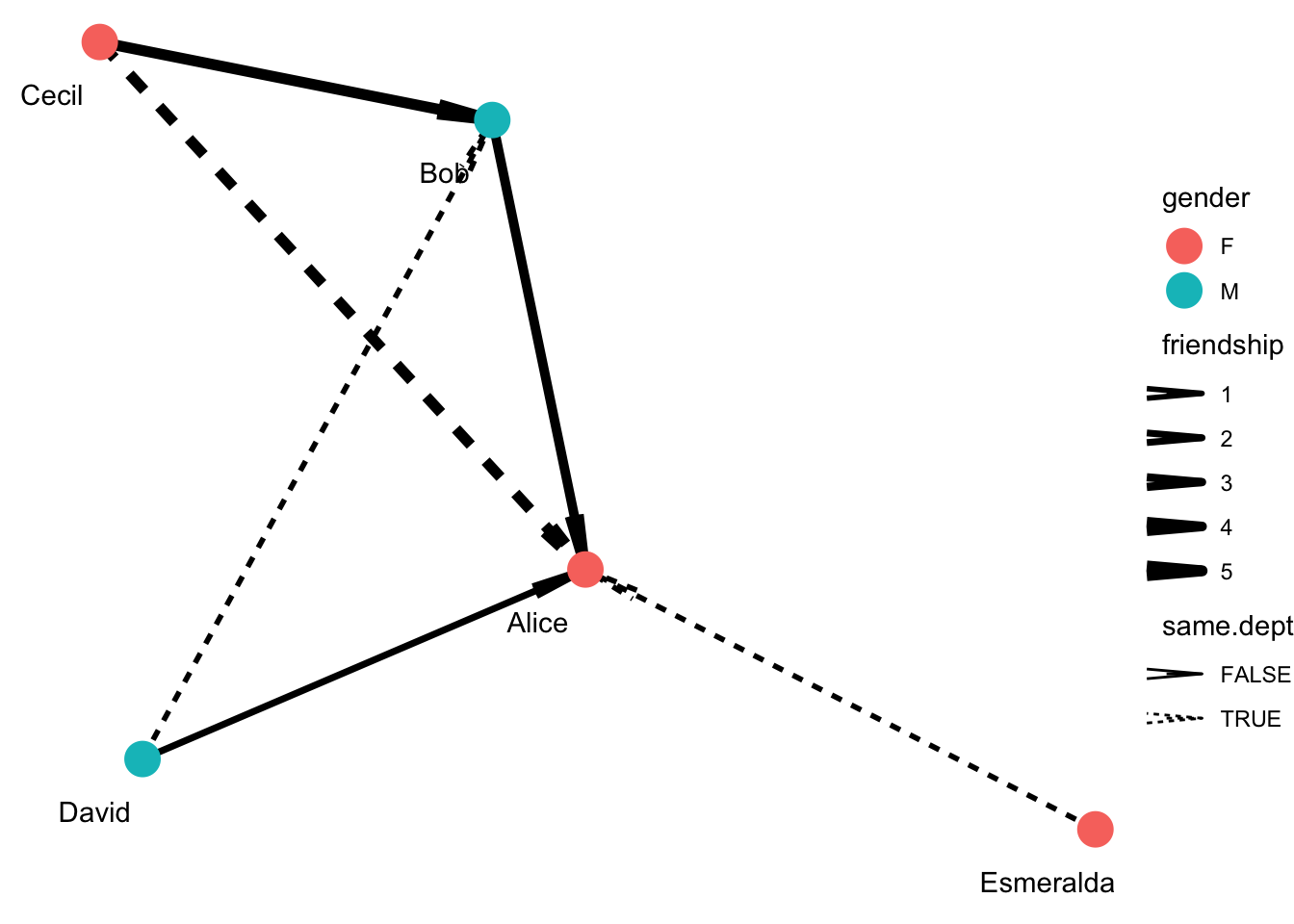
Facet
One big advantage of {ggraph} is to use facet. It can be facet_edges or facet_nodes or facet_graph. Here I will only show example of facet_nodes.
Code
g = g %>% set_vertex_attr("dept",value=c("sale","IT","sale","IT","sale")) %>%
set_edge_attr("same.dept",value=c(F,F,T,F,T,T))
# facet based on the dept
ggraph(g, layout="kk") +
facet_nodes(~dept, drop = F) +
geom_edge_link(aes(edge_width=friendship, linetype=same.dept), arrow = arrow(angle=5, length = unit(0.3, "inches"))) +
geom_node_point(aes(color=gender), size=6) +
geom_node_text(aes(label=name), nudge_y = -0.1, nudge_x = -0.1) +
scale_edge_width(range = c(1, 2))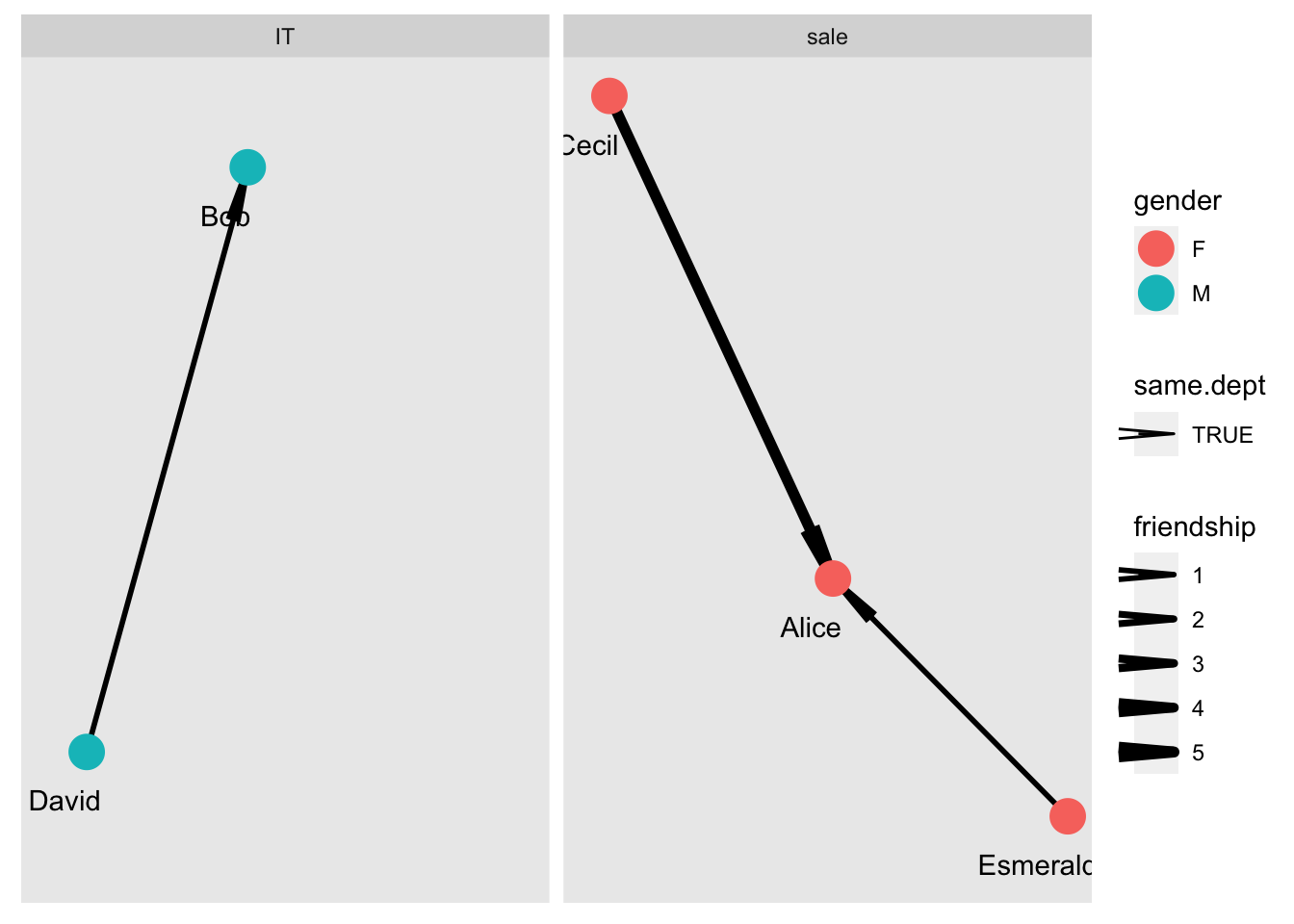
Layout
There are many layouts available for both igraph and ggraph pacakges. Igraph provides a huge amount of layouts. https://igraph.org/r/doc/layout_.html
- Standard layouts
- bipartite: minimize edge-crossings in a simple two-row (or column) layout for bipartite graphs.
- star: place one node in the center and the rest equidistantly around it.
as_star() - circle: place nodes in a circle in the order of their index. Consider using layout_igraph_linear with circular=TRUE for more control.
in_circle() - nicely: default, tries to pick an appropriate layout.
nicely - dh: uses Davidson and Harels simulated annealing algorithm to place nodes.
with_dh() - gem: place nodes on the plane using the GEM force-directed layout algorithm.
with_gem - graphopt: uses the Graphopt algorithm based on alternating attraction and repulsion to place nodes.
with_graphopt() - grid:place nodes on a rectangular grid.
on_grid() - mds: perform a multidimensional scaling of nodes using either the shortest path or a user supplied distance.
with_mds() - sphere: place nodes uniformly on a sphere - less relevant for 2D visualizations of networks.
with_sphere() - randomly: places nodes uniformly random.
randomly - fr: places nodes according to the force-directed algorithm of Fruchterman and Reingold.
with_fr() - kk: uses the spring-based algorithm by Kamada and Kawai to place nodes.
with_kk() - drl: uses the force directed algorithm from the DrL toolbox to place nodes.
with_drl() - lgl: uses the algorithm from Large Graph Layout to place nodes. See with_lgl
with_lgl()
- Hierarchical layouts
- tree: uses the Reingold-Tilford algorithm to place the nodes below their parent with the parent centered above its children.
as_tree() - sugiyama: designed for directed acyclic graphs (that is, hierarchies where multiple parents are allowed) it minimizes the number of crossing edges.
- tree: uses the Reingold-Tilford algorithm to place the nodes below their parent with the parent centered above its children.
Here we are going to show an example how to switch standard layout using the same data
Code
par(mfrow=c(2,3))
# star layout -- help determine center
coords <- layout_(g, as_star())
plot(g, layout = coords, edge.arrow.size=0.4)
title("start")
# circle layout
coords <- layout_(g, in_circle())
plot(g, layout = coords, edge.arrow.size=0.4)
title("circle")
# grid
coords <- layout_(g, on_grid())
plot(g, layout = coords, edge.arrow.size=0.4)
title("grid")
# nicely
coords <- layout_(g, nicely())
plot(g, layout = coords, edge.arrow.size=0.4)
title("nicely")
# kk
coords <- layout_(g, with_kk())
plot(g, layout = coords, edge.arrow.size=0.4)
title("Kamada and Kawai(kk)")
# fr
coords <- layout_(g, with_fr())
plot(g, layout = coords, edge.arrow.size=0.4)
title("force-directed(fr)")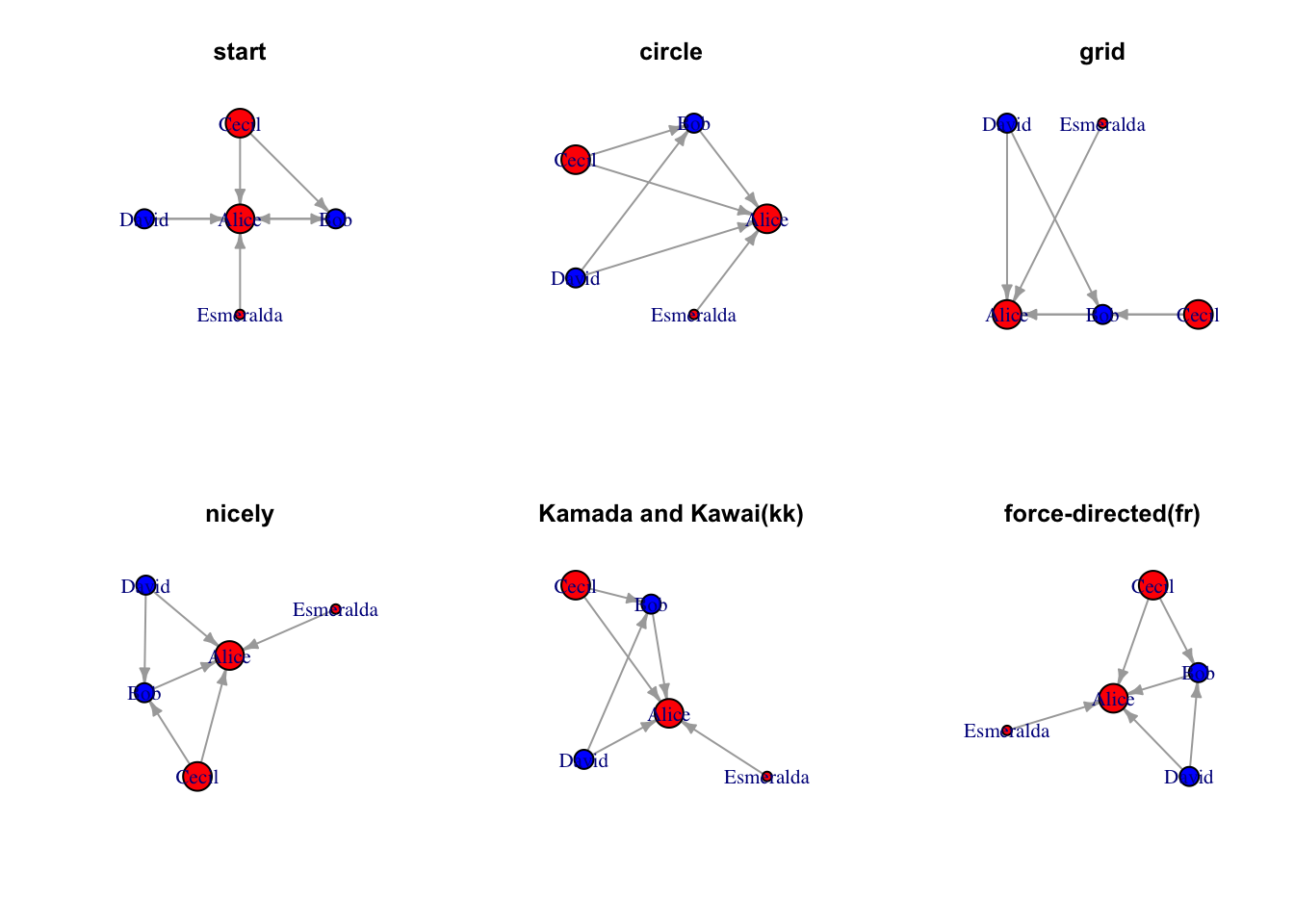
Hierarchical layouts can plot data in layer. Here show example how to use sugiyama layout
Code
# make different dept nodes at different node
g = g %>% set_vertex_attr("dept",value=c("sale","IT","sale","IT","sale")) %>%
set_edge_attr("same.dept",value=c(F,F,T,F,T,T))
v = as_data_frame(g, "vertices") %>% as_tibble %>%
mutate(layer=ifelse(dept=="sale",1,2))
e = as_data_frame(g, what="edges") %>% as_tibble %>%
mutate(width=friendship) %>%
mutate(lty=ifelse(same.dept,1,2))
g = g %>% set_edge_attr("width",value=e$width) %>% set_edge_attr("lty",value=e$lty)
lay1 <- layout_with_sugiyama(g, layers=v$layer, attributes="all")
plot(lay1$extd_graph, edge.curved=T)
legend("topleft", legend=unique(v$gender),pch=21,pt.bg=c("red","blue"), title="gender", box.lty=0)
legend("left",legend=unique(e$same.dept),lty=c(1,2), title = "same.dept",box.lty=0)
legend("topright", legend=sort(unique(e$friendship)), lwd=sort(unique(e$friendship)), title="friendship", box.lty=0)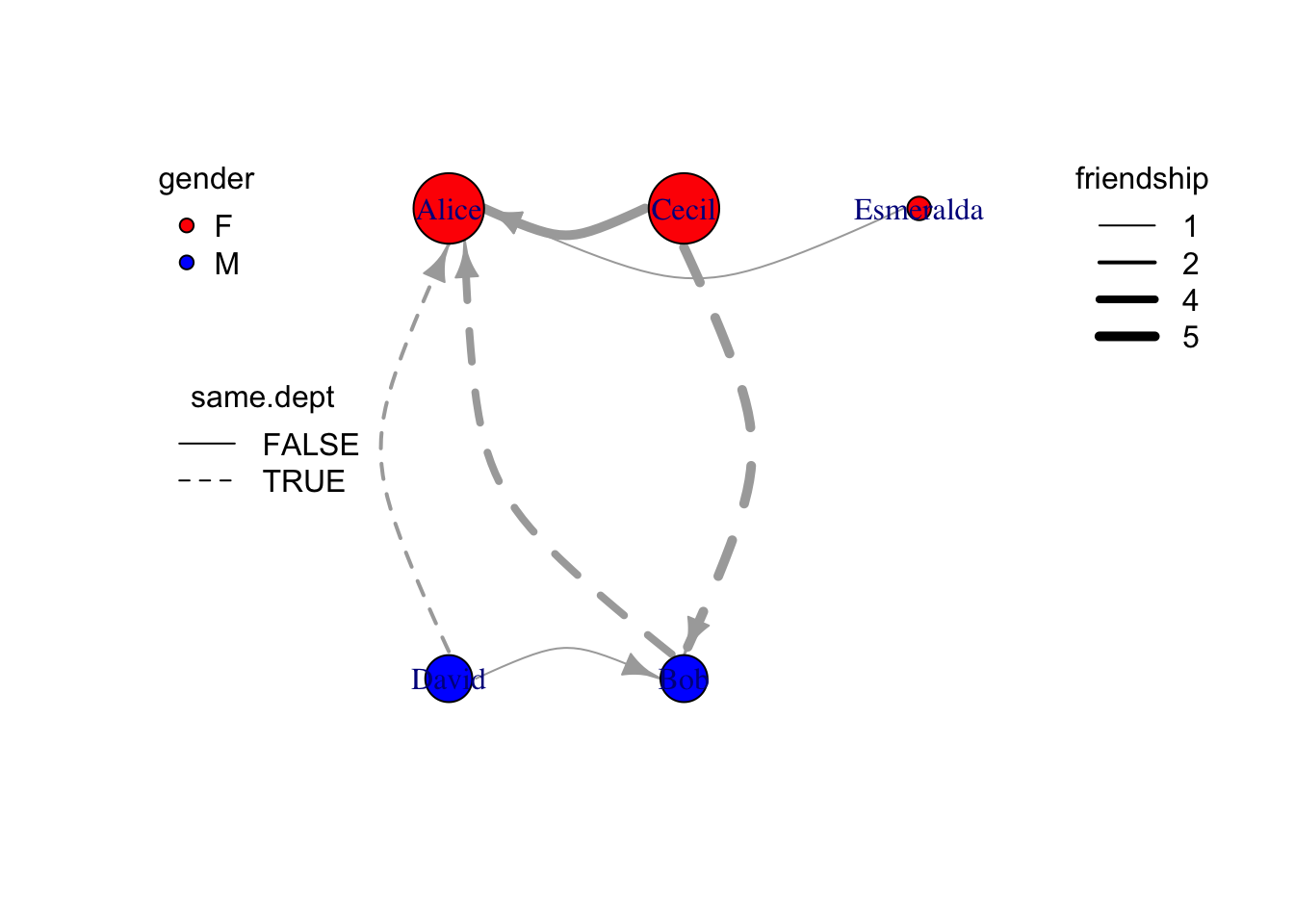
ggraph can use all the layout mentioned above by specifying it in ggraph(g, layout=...). Besides, ggraph has addtional useful layout.
- dendrogram: dendrogram layout not only take in graph object but also dendrogram object (
as.dendrogram(hclust(dist(...)))). ggraph will automatically convert dendrogram to igraph byden_to_igraph. It ususally plots usinggeom_edge_diagonal()orgeom_edge_elbow()
Code
den <- as.dendrogram(hclust(dist(mtcars)))
p1 = ggraph(den, 'dendrogram') +
geom_edge_diagonal() +
geom_node_text(aes(label=label), angle=90, nudge_y=-30, size=3) +
theme_void()
p2 = ggraph(den, 'dendrogram', circular = TRUE) +
geom_edge_elbow() +
geom_node_text(aes(label=label), angle=45, size=2) +
coord_fixed()+
theme_void()
grid.arrange(p1,p2,ncol=2)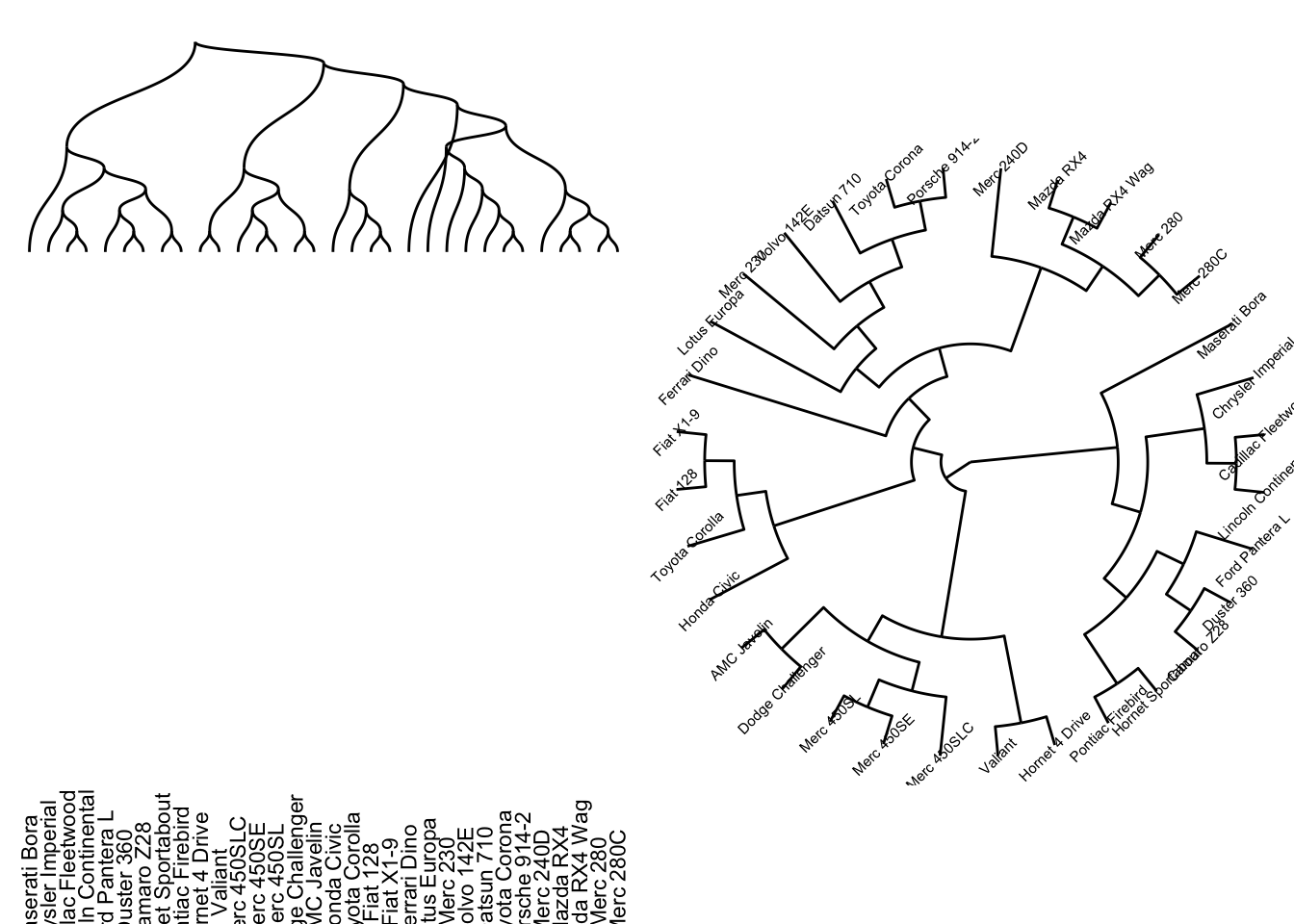
- hive: make nodes group into a axis and connecting axis instead.
Code
V(g)$age_range = factor(V(g)$age_range)
ggraph(g, 'hive', axis = age_range, sort.by = age) +
geom_edge_hive(aes(color = factor(same.dept), edge_width=friendship)) +
geom_axis_hive(aes(color = age_range), size = 3, label = FALSE) +
coord_fixed() +
scale_edge_width(range=c(1,3))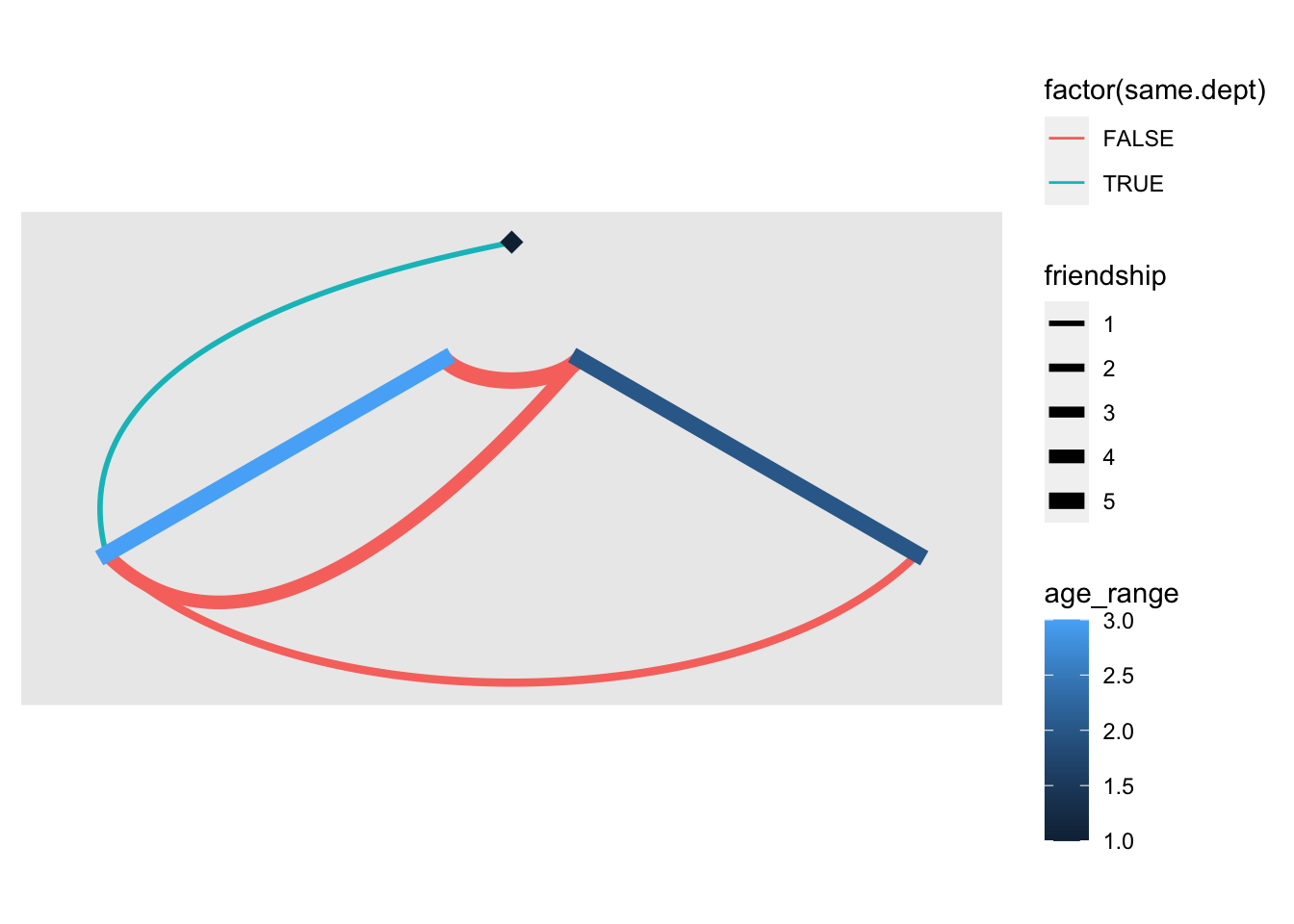
- linear: make nodes only the same line so that arc connections were made
Code
ggraph(g, layout = 'linear', sort.by = age) +
geom_edge_arc(aes(colour = factor(same.dept), edge_alpha=friendship)) +
geom_node_point(aes(color=gender), size=4, alpha=0.5) +
geom_node_text(aes(label=name), angle=45) +
theme_void() +
scale_edge_alpha(range=c(0.3,1))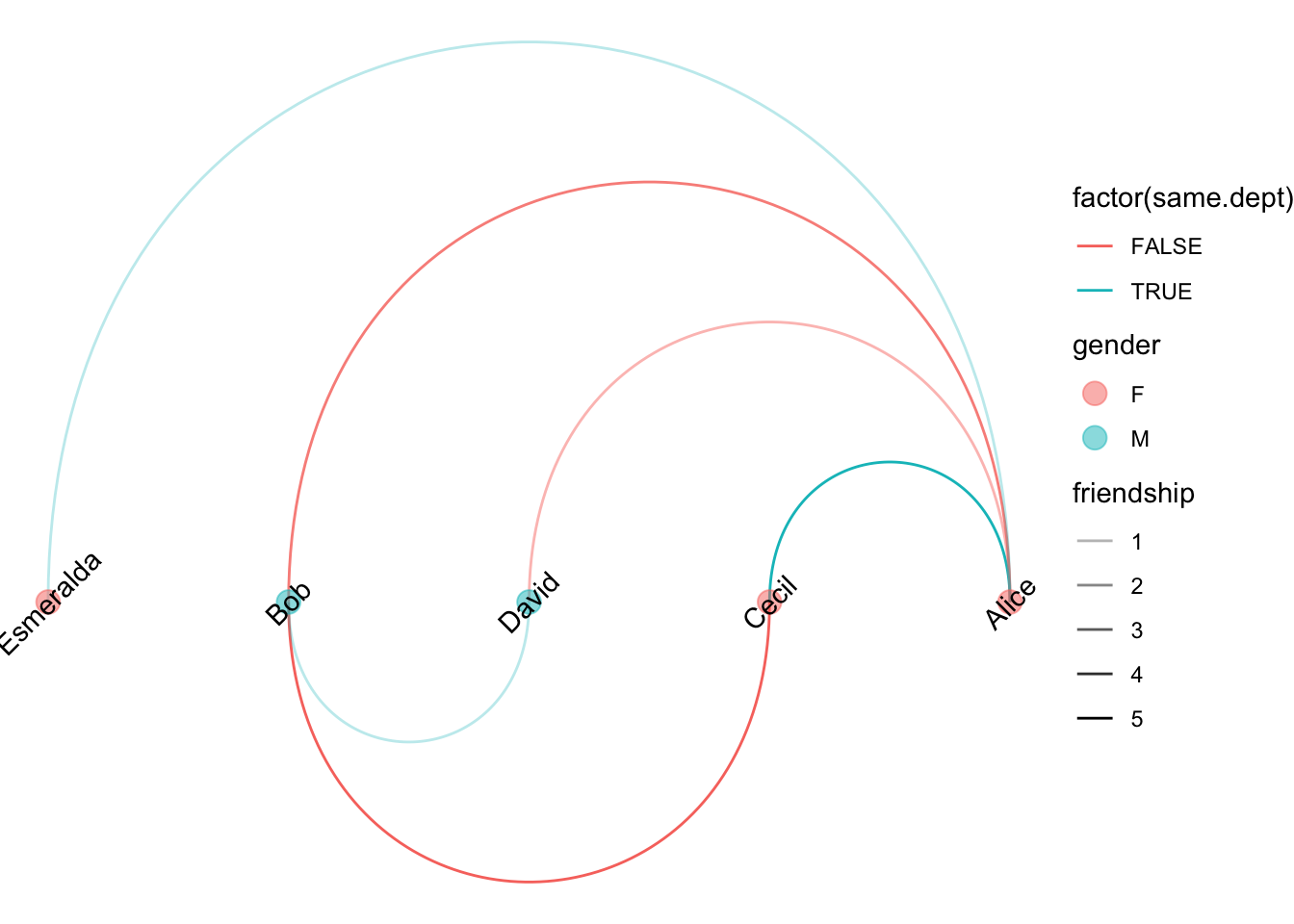
More functions about ggraph refer to https://www.rdocumentation.org/packages/ggraph/versions/1.0.2
other packages for graph visualization
There are many other packages available for graph visualization and network analysis. In this series, I will only list the link here for the further reference. I may come back to further this topic in the future when necessary.
Network analysis tool: Statnet1
Network visualization: ggnet2
Interactive network :